- Cyclase-associated protein family
-
CAP N-terminal 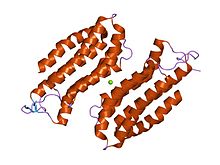
structure of the n-terminal domain of the adenylyl cyclase-associated protein (cap) from dictyostelium discoideum. Identifiers Symbol CAP_N Pfam PF01213 InterPro IPR013992 PROSITE PDOC00835 SCOP 1s0p Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary CAP C-terminal 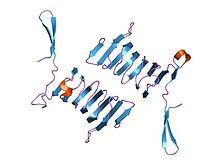
c-terminal domain of cyclase associated protein with pro 505 replaced by ser (p505s) Identifiers Symbol CAP_C Pfam PF08603 Pfam clan CL0391 InterPro IPR013912 PROSITE PDOC00835 SCOP 1kq5 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary In molecular biology, the cyclase-associated protein family (CAP) is a family of highly conserved actin-binding proteins present in a wide range of organisms including yeast, flies, plants, and mammals. CAPs are multifunctional proteins that contain several structural domains. CAP is involved in species-specific signalling pathways.[1][2][3][4] In Drosophila, CAP functions in Hedgehog-mediated eye development and in establishing oocyte polarity. In Dictyostelium (slime mould), CAP is involved in microfilament reorganisation near the plasma membrane in a PIP2-regulated manner and is required to perpetuate the cAMP relay signal to organise fruitbody formation. In plants, CAP is involved in plant signalling pathways required for co-ordinated organ expansion. In yeast, CAP is involved in adenylate cyclase activation, as well as in vesicle trafficking and endocytosis. In both yeast and mammals, CAPs appear to be involved in recycling G-actin monomers from ADF/cofilins for subsequent rounds of filament assembly.[5][6] In mammals, there are two different CAPs (CAP1 and CAP2) that share 64% amino acid identity.
All CAPs appear to contain a C-terminal actin-binding domain that regulates actin remodelling in response to cellular signals and is required for normal cellular morphology, cell division, growth and locomotion in eukaryotes. CAP directly regulates actin filament dynamics and has been implicated in a number of complex developmental and morphological processes, including mRNA localisation and the establishment of cell polarity. Actin exists both as globular (G) (monomeric) actin subunits and assembled into filamentous (F) actin. In cells, actin cycles between these two forms. Proteins that bind F-actin often regulate F-actin assembly and its interaction with other proteins, while proteins that interact with G-actin often control the availability of unpolymerised actin. CAPs bind G-actin.
In addition to actin-binding, CAPs can have additional roles, and may act as bifunctional proteins. In Saccharomyces cerevisiae (Baker's yeast), CAP is a component of the adenylyl cyclase complex (Cyr1p) that serves as an effector of Ras during normal cell signalling. S. cerevisiae CAP functions to expose adenylate cyclase binding sites to Ras, thereby enabling adenylate cyclase to be activated by Ras regulatory signals. In Schizosaccharomyces pombe (Fission yeast), CAP is also required for adenylate cyclase activity, but not through the Ras pathway. In both organisms, the N-terminal domain is responsible for adenylate cyclase activation, but the S. cerevisiae and S. pombe N-termini cannot complement one another. Yeast CAPs are unique among the CAP family of proteins, because they are the only ones to directly interact with and activate adenylate cyclase.[7] S. cerevisiae CAP has four major domains. In addition to the N-terminal adenylate cyclase-interacting domain, and the C-terminal actin-binding domain, it possesses two other domains: a proline-rich domain that interacts with Src homology 3 (SH3) domains of specific proteins, and a domain that is responsible for CAP oligomerisation to form multimeric complexes (although oligomerisation appears to involve the N- and C-terminal domains as well). The proline-rich domain interacts with profilin, a protein that catalyses nucleotide exchange on G-actin monomers and promotes addition to barbed ends of filamentous F-actin.[5] Since CAP can bind profilin via a proline-rich domain, and G-actin via a C-terminal domain, it has been suggested that a ternary G-actin/CAP/profilin complex could be formed.
The N-terminal domain has an all-alpha structure consisting of six helices in a bundle with a left-handed twist and an up-and-down topology.[8]
The C-terminal domain is responsible for G-actin-binding. This domain has a superhelical structure, where the superhelix turns are made of two beta-strands each.[9]
References
- ^ Hubberstey AV, Mottillo EP (April 2002). "Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization". FASEB J. 16 (6): 487–99. PMID 11919151.
- ^ Deeks MJ, Rodrigues C, Dimmock S, Ketelaar T, Maciver SK, Malhó R, Hussey PJ (August 2007). "Arabidopsis CAP1 - a key regulator of actin organisation and development". J. Cell. Sci. 120 (Pt 15): 2609–18. doi:10.1242/jcs.007302. PMID 17635992.
- ^ Freeman NL, Field J (February 2000). "Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly". Cell Motil. Cytoskeleton 45 (2): 106–20. doi:10.1002/(SICI)1097-0169(200002)45:2<106::AID-CM3>3.0.CO;2-3. PMID 10658207.
- ^ Hofmann A, Hess S, Noegel AA, Schleicher M, Wlodawer A (October 2002). "Crystallization of cyclase-associated protein from Dictyostelium discoideum". Acta Crystallogr. D Biol. Crystallogr. 58 (Pt 10 Pt 2): 1858–61. doi:10.1107/S0907444902013306. PMID 12351838.
- ^ a b Bertling E, Quintero-Monzon O, Mattila PK, Goode BL, Lappalainen P (April 2007). "Mechanism and biological role of profilin-Srv2/CAP interaction". J. Cell. Sci. 120 (Pt 7): 1225–34. doi:10.1242/jcs.000158. PMID 17376963.
- ^ Bertling E, Hotulainen P, Mattila PK, Matilainen T, Salminen M, Lappalainen P (May 2004). "Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells". Mol. Biol. Cell 15 (5): 2324–34. doi:10.1091/mbc.E04-01-0048. PMC 404026. PMID 15004221. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=404026.
- ^ Shima F, Okada T, Kido M, Sen H, Tanaka Y, Tamada M, Hu CD, Yamawaki-Kataoka Y, Kariya K, Kataoka T (January 2000). "Association of yeast adenylyl cyclase with cyclase-associated protein CAP forms a second Ras-binding site which mediates its Ras-dependent activation". Mol. Cell. Biol. 20 (1): 26–33. PMC 85033. PMID 10594005. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=85033.
- ^ Ksiazek D, Brandstetter H, Israel L, Bourenkov GP, Katchalova G, Janssen KP, Bartunik HD, Noegel AA, Schleicher M, Holak TA (September 2003). "Structure of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum". Structure 11 (9): 1171–8. doi:10.1016/S0969-2126(03)00180-1. PMID 12962635.
- ^ Dodatko T, Fedorov AA, Grynberg M, Patskovsky Y, Rozwarski DA, Jaroszewski L, Aronoff-Spencer E, Kondraskina E, Irving T, Godzik A, Almo SC (August 2004). "Crystal structure of the actin binding domain of the cyclase-associated protein". Biochemistry 43 (33): 10628–41. doi:10.1021/bi049071r. PMID 15311924.
This article includes text from the public domain Pfam and InterPro IPR013992
This article includes text from the public domain Pfam and InterPro IPR013912
Categories:- Protein families
Wikimedia Foundation. 2010.
