- Meiotic recombination checkpoint
-
The meiotic recombination checkpoint monitors the meiotic recombination during meiosis, and blocks the entry into metaphase I if the recombination is not efficiently processed. Meitotic recombination contributes to the cells in two different ways. First, to achieve proper segregation, each pair of homologous chromosomes must be linked to each other to maintain a certain level of tension between them. Such tension is supposed to help the assembly of spindles and generally depends on meitotic recombination. Secondly, Meitotic recombination increases the genetic diversity of gametes, making them readily adapt to new environment.
Contents
Overview
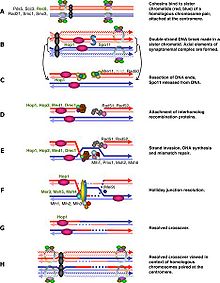
Spo11 catalyzes a double strand break in one of the two homologous chromosomes to induce meiotic recombination. DSB-dependent meiotic recombination checkpoint monitors the repair of these DSBs while DSB-independent meiotic recombination checkpoint examines the asynapsis of each homolog pair which is the consequence of uncompleted DSB induction.
Generally speaking, the cell cycle regulation of meiosis is similar to that of mitosis. As in the mitotic cycle, these transitions are regulated by combinations of gene regulatory factors, cyclin-Cdk complex and the APC.[1] The first major regulatory transition occurs in late G1, when the Start of meiotic cycle is activated by Ime1 instead of Cln3/Cdk1 in mitosis. The second major transition occurs at the entry into metaphase I. The main purpose of this step is to make sure that DNA replication has completed without error so that spindle pole bodies can separate. The event is triggered by the activation of M-Cdk in late prophase I. Then the spindle assembly checkpoint examines the attachment of microtubules at kinetochores and APCCdc20 can initiate the metaphase I. The special chromosome separation in meiosis, homologous chromosomes separation in meiosis I and chromatids separation in meiosis II, requires special tension between homologous chromatids and non-homologous chromatids for distinguishing microtubule attachment and it relies on the programmed DNA double strand break (DSB) and repair in prophase I. Therefore meiotic recombination checkpoint can be a kind of DNA damage response at specific time spot. On the other hand, the meiotic recombination checkpoint also makes sure that meiotic recombination does happen in every pair of homologs.
DSB-dependent pathway
The abrupt onset of M-Cdk in late prophase I depends on the positive transcription regulation feedback loop consisting of Ime2, Ndt80 and Cdk/cyclin complex. However the activation of M-Cdk is controlled by the genereral phosphorylation switch Wee1/Cdc25. Wee1 activity is high in early prophase I and the accumulation of Cdc25 activates M-Cdk by direct phosphorylation and marking Wee1 to be degraded. Meiotic recombination begins with a double strand break induced by Spo11,[2] and these DSBs must be repaired before metaphase I. The cell monitor these DSBs via ATM pathway, in which Cdc25 is suppressed when DSB lesion is detected. This pathway is the same as classical DNA damage response and is the part we know the best in meiotic recombination checkpoint.
DSB-independent pathway
The DSB-independent pathway was proposed when people studied spo11 mutant cells in some species and found that these Spo11 cells could not process to metaphase I even in the absence of DSB.[3] The direct purpose of these DSBs is to help with the condensation of chromosomes. Even though the initial homolog paring in early leptotene is just random interactions, the further progression into presynaptic alignment depends on the formation of double strand breaks and single strand transfer complexes.[1][4] Therefore the unsynapsed chromosomes in Spo11 cells can be a target of checkpoint. An AAA–adenosine triphosphatase (AAA-ATPase) was found to be essential in this pathway.[5] but the mechanism is not yet clear. Some other studies also drew sex body formation into attention, and the signaling could be either structure based or transcription regulation such as meiotic sex chromosome inactivation.[6][7] Under this cascade, failure to synapse will maintain the gene expression from sex chromosomes and some products may inhibit cell cycle progression. meiotic sex chromosome inactivation only happens in male, that may partially be the reason why only Spo11 mutant spermatocytes but not oocytes fail to transit from prophase I to metaphase I.[3][8] However the asynapsis does not happen only within sex chromosomes, and such trancription regulation was suspended until it was further expanded to all the chromosomes as meiotic silencing of unsynapsed chromatin,[9] but the effector gene is not found yet.
References
- ^ a b Morgan, D (2007). "Chapter 9: Meitosis". The Cell Cycle: Principles of Control. London: New Science Press Ltd. ISBN 0-87893-508-8.
- ^ Malik, S-B; Pightling, AW; Stefaniak, LM; Schurko, AM; Logsdon Jr, JM; Logsdon, John M. (August 2008). Hahn, Matthew W.. ed. "An Expanded Inventory of Conserved Meiotic Genes Provides Evidence for Sex in Trichomonas vaginalis". PLoS ONE 3 (8): e2879. doi:10.1371/journal.pone.0002879. PMC 2488364. PMID 18663385. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2488364.
- ^ a b Barchi, M; Mahadevaiah, S; Di Giacomo, M; Baudat, F; De Rooij, DG; Burgoyne, PS; Jasin, M; Keeney, S (August 2005). "Surveillance of Different Recombination Defects in Mouse Spermatocytes Yields Distinct Responses despite Elimination at an Identical Developmental Stage". Molecular and Cellular Biology 25 (16): 7203–15. doi:10.1128/MCB.25.16.7203-7215.2005. PMC 1190256. PMID 16055729. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1190256.
- ^ Storlazzi, A; Tessé, S; Gargano, S; James, F; Kleckner, N; Zickler, D (November 2003). "Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division". Genes & Development 17 (21): 2675–87. doi:10.1101/gad.275203. PMC 280617. PMID 14563680. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=280617.
- ^ Bhalla, N; Dernburg, AF (December 2005). "A Conserved Checkpoint Monitors Meiotic Chromosome Synapsis in Caenorhabditis elegans". Science 310 (5754): 1683–6. doi:10.1126/science.1117468. PMID 16339446.
- ^ Odorisio, T; Rodriguez, TA; Evans, EP; Clarke, AR; Burgoyne, PS (1998). "The meiotic checkpoint monitoring sypapsis eliminates spermatocytes via p53-independent apoptosis". Nature Genetics 18 (3): 257–61. doi:10.1038/ng0398-257. PMID 9500548.
- ^ Turner, J; Mahadevaiah, SK; Elliott, DJ; Garchon, HJ; Pehrson, JR; Jaenisch, R; Burgoyne, PS (2002). "Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist". Journal of Cell Science 115 (Pt 21): 4097–105. doi:10.1242/jcs.00111. PMID 12356914.
- ^ Di Giacomo, M; Barchi, M; Baudat, F; Edelmann, W; Keeney, S; Jasin, M (2005). "Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants". Proc. Natl. Acad. Sci. 102 (3): 737–42. doi:10.1073/pnas.0406212102. PMC 545532. PMID 15640358. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=545532.
- ^ Manterola, M; Page, J; Vasco, C; Berríos, S; Parra, MT; Viera, A; Rufas, JS; Zuccotti, M et al. (2009). Hawley, R. Scott. ed. "A High Incidence of Meiotic Silencing of Unsynapsed Chromatin Is Not Associated with Substantial Pachytene Loss in Heterozygous Male Mice Carrying Multiple Simple Robertsonian Translocations". PLoS Genet 5 (8): e1000625. doi:10.1371/journal.pgen.1000625. PMC 2726437. PMID 19714216. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2726437.
Categories:
Wikimedia Foundation. 2010.