- Multiregional origin of modern humans
-
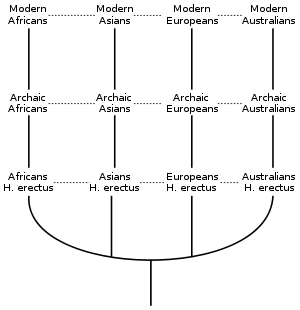 A graph detailing the evolution to modern humans using the Multiregional theory of human evolution. The horizontal lines represent 'multiregional evolution' gene flow between regional lineages. In Weidenreich's original graphic (which is more accurate than this one), there were also diagonal lines between the populations, e.g. between African H. erectus and Archaic Asians and between Asian H. erectus and Archaic Africans. This created a "trellis" (as Wolpoff called it) or a "network" that emphasized gene flow between geographic regions and within time. It is important to remember that the populations on the chart are not discrete - i.e., they do not represent different species, but are samples within a long lineage experiencing extensive gene flow.
A graph detailing the evolution to modern humans using the Multiregional theory of human evolution. The horizontal lines represent 'multiregional evolution' gene flow between regional lineages. In Weidenreich's original graphic (which is more accurate than this one), there were also diagonal lines between the populations, e.g. between African H. erectus and Archaic Asians and between Asian H. erectus and Archaic Africans. This created a "trellis" (as Wolpoff called it) or a "network" that emphasized gene flow between geographic regions and within time. It is important to remember that the populations on the chart are not discrete - i.e., they do not represent different species, but are samples within a long lineage experiencing extensive gene flow.
The multiregional hypothesis is a scientific model that provides an explanation for the pattern of human evolution. The hypothesis holds that humans first arose near the beginning of the Pleistocene two million years ago and subsequent human evolution has been within a single, continuous human species. This species encompasses archaic human forms such as Homo erectus and Neanderthals as well as modern forms, and evolved worldwide to the diverse populations of modern Homo sapiens sapiens. The theory contends that humans evolve through a combination of adaptation within various regions of the world and gene flow between those regions. Proponents of multiregional origin point to fossil and genomic data and continuity of archaeological cultures as support for their hypothesis. [1]
A majority of scholars prefer the primary competing hypothesis, which postulates a recent African origin of modern humans, contending that modern humans arose in Africa, most likely around 150-200,000 years ago, moving out of Africa around 50-60,000 years ago to replace archaic human forms.[2]
Contents
Overview
The idea of multiregional evolution goes back to Franz Weidenreich in the early 20th century, who saw similarities between the archaic human Peking man fossils and modern humans in China.[3][4] Weidenreich merged the Asia hypothesis into the multiregional, extending many other regions into his theory from the Old World with gene flow between various populations which influenced many anthropologists of the time.[5][6]
The term "multiregional hypothesis" was first applied to the theory in the early 1980s by Milford H. Wolpoff and colleagues, who used the theory to explain these and other similarities between archaic humans and modern humans in various regions, in what they called regional continuity.[7][8]
Wolpoff rejected the proposal by Coon of parallel evolution,[8] and proposed that the mechanism of clinal variation allowed for the necessary balance between both local selection and overall evolution as a global species, with Homo erectus, Neanderthals, Homo sapiens and other human forms as subspecies. This species arose in Africa two million years ago as H. erectus and then spread out over the world, developing adaptations to regional conditions. Some populations became isolated for periods of time, developing in different directions, but through continuous interbreeding, replacement, genetic drift and selection, adaptations that were an advantage anywhere on earth would spread, keeping the development of the species in the same overall direction while maintaining adaptations to regional factors. By these mechanisms, surviving local varieties of the species evolved into modern humans, retaining some regional adaptations but with many features common to all regions.[8]
Fossil evidence
Proponents of the multiregional hypothesis see regional continuity of certain morphological traits from archaic humans to modern humans, demonstrating regional genetic continuity, even as changes in other traits occur in parallel over time across all regions, demonstrating lateral genetic exchange.[9] For example, in 2001 Wolpoff and colleagues published an analysis of character traits of the skulls of early modern human fossils in Australia and central Europe. They concluded that the diversity of these recent humans could not "result exclusively from a single late Pleistocene dispersal", and implied dual ancestry from Javan Homo erectus for Australia and from Neanderthals for Central Europe.[10][11]
Southeast Asia
Alan Thorne held that there was regional continuity in the human fossils in southeast Asia. Wolpoff, initially skeptical, became convinced when reconstructing the Sangiran 17 Homo erectus skull from Indonesia, when he was surprised that the skull's face to vault angle matched that of the Australian modern human Kow Swamp 1 skull. Wolpoff had expected the skull to match that of the Homo erectus specimens from China like the Dali skull, but instead, the face to vault angle seemed to be retained regionally over time, even while the fossils in the two regions showed parallel increases in brain case size and parallel reductions in masticatory structures over the intervening approximately 750,000 years.[9]
China
Franz Weidenreich, who oversaw the excavations of numerous "Peking man" Homo erectus fossils at Zhoukoudian in the early 20th century, believed the fossil record demonstrated certain unique features linking prehistoric and modern human populations in China. Many subsequent Chinese paleoanthropologists, such as Wu Xinzhi, were also disposed to favor the multiregional hypothesis for the same reason.[4]
More recent finds provide additional support for regional human continuity in China. The Tianyuan 1 specimen unearthed in 2003 in Tianyuan Cave, Zhoukoudian, and Carbon 14 dated to 42-39 kya exhibits a series of typical modern human features such as a distinct chin. However, the skeleton also has archaic traits such as low anterior to posterior dental proportions indicating relatively large molars and certain leg bone proportions typical of archaic forms such as Neanderthals. Shang et al. (1999) conclude that this combination of modern and archaic traits "implies that a simple spread of modern humans from Africa is unlikely."[12] A jaw bone found in 2008 and dated to 110 kya may also exhibit a mixture of archaic and modern human traits.[13]
Europe
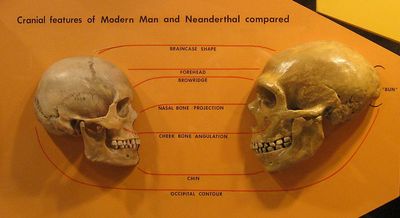
Proponents of the multiregional hypothesis argue for regional continuity in Europe on the basis of skeletal anatomy, morphology and genetics of speech, and the archaeology of the middle to upper paleolithic transition, which they believe to be inconsistent with the possibility of complete replacement of the Neanderthals in Europe without interbreeding.[14]
Some detractors of the theory have argued, in contrast, that the morphological differences between Neanderthals and early and modern humans indicate that they are different species, based on skull differences more distinct than between any subspecies pairs examined except for the two subspecies of gorilla, implying limited or no interbreeding.[15][16]
Many of the multiregional claims regarding skeletal morphology in Europe center on forms with both archaic Neanderthal traits and modern traits, to provide evidence of interbreeding rather than replacement. Examples include the Lapedo child found in Portugal[17] and the Oase 1 mandible from Peştera cu Oase, Romania,[18] though the Lapedo child example is disputed by some.[19] In a 2007 paper examining numerous samples from European early modern humans, later European humans from the Gravettian period, and the earlier Neanderthal and east African populations from whom the first two populations could have descended, Erik Trinkaus identified numerous features in the later European samples which were absent from the African sample, but present in the Neanderthal sample. These features included various aspects of skull and mandible shape, tooth shape and size, and shapes and proportions of other bones. Trinkaus concluded that early modern Europeans had predominant African ancestry with a substantial degree of admixture from the Neanderthals then indigenous to Europe.[20][21]
Genetic evidence
Genetic evidence from the late 1980s on the mitochondrial genome indicated that all living women can trace their maternal line of descent to a single female living in Africa about 200,000 years ago, the so-called Mitochondrial Eve. This led to a hypothesis that Homo sapiens evolved in Africa, with a small founder population of humans leaving Africa and eventually replacing all archaic humans then living elsewhere without interbreeding.[citation needed] Recent analyses of DNA taken directly from Neanderthal specimens, however, indicates that they contributed to the genome of all humans outside of Africa. The Homo sapiens who populated the world outside Africa all have Neanderthals among their descendents. Denisova hominins also contributed to the DNA of Milanesians and Australians.
Mitochondrial DNA
A 1987 analysis of mitochondrial DNA from 147 people from around the world indicated that their mitochondrial lineages all coalesced in a common ancestor in Africa about 200,000 years ago. The analysis suggested that this reflected the worldwide expansion of modern humans as a new species, replacing rather than mixing with local archaic humans.[22] Later analysis of mitochondrial DNA from neanderthals and from the denisova hominin indicated that those mitochondrial strains had diverged from the living human mitochondrial line long before 200,000 years ago, consistent with lack of interbreeding between early modern and archaic humans.[23][24]
The original mitochondrial DNA results and the resulting recent African replacement theory posed a serious challenge to the multiregional hypothesis.[25] Mitochondrial DNA alone, however, could not entirely rule out interbreeding between early modern and archaic humans, since archaic human mitochondrial strains from such interbreeding could have been lost due to genetic drift or a selective sweep.[26][27]
Nuclear DNA
Initial analysis of Y chromosome DNA, which like mitochondrial DNA is inherited from only one parent, was consistent with a recent African replacement model. However, the mitochondrial and Y chromosome data could not be explained by the same modern human expansion out of Africa; the Y chromosome expansion would have involved genetic mixing that retained regionally local mitochondrial lines. In addition, the Y chromosome data indicated a later expansion back into Africa from Asia, demonstrating that gene flow between regions was not unidirectional.[28]
An early analysis of 15 noncoding sites on the X chromosome found additional inconsistencies with the recent African replacement hypothesis. The analysis found a multimodal distribution of coalescence times to the most recent common ancestor for those sites, contrary to the predictions for recent African replacement; in particular, there were more coalescence times near 2 million years ago (mya) than expected, suggesting an ancient population split around the time humans first emerged from Africa as Homo erectus, rather than more recently as suggested by the mitochondrial data. While most of these X chromosome sites showed greater diversity in Africa, consistent with African origins, a few of the sites showed greater diversity in Asia rather than Africa. For four of the 15 gene sites that did show greater diversity in Africa, the sites' varying diversity by region could not be explained by simple expansion from Africa, as would be required by the recent African replacement hypothesis.[29]
Later analyses of X chromosome and autosomal DNA continued to find sites with deep coalescence times inconsistent with a single origin of modern humans,[30][31][32][33][34][35] diversity patterns inconsistent with a recent expansion from Africa,[36] or both.[37][38] For example, analyses of a region of RRM2P4 (ribonucleotide reductase M2 subunit pseudogene 4) showed a coalescence time of about 2 Mya, with a clear root in Asia,[39][40] while the MAPT locus at 17q21.31 is split into two deep genetic lineages, one of which is common in and largely confined to the present European population, suggesting inheritance from Neanderthals.[41][42][43][44] In the case of the Microcephalin D allele, evidence for rapid recent expansion indicated introgression from an archaic population.[45][46][47][48]
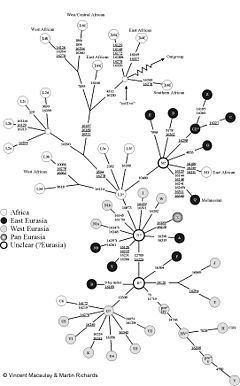
In a 2005 review and analysis of the genetic lineages of 25 chromosomal regions, Alan Templeton found evidence of more than 34 occurrences of gene flow between Africa and Eurasia. Of these occurrences, 19 were associated with continuous restricted gene exchange through at least 1.46 million years ago; only 5 were associated with a recent expansion from Africa to Eurasia. Three were associated with the original expansion of Homo erectus out of Africa around 2 million years ago, 7 with an intermediate expansion out of Africa at a date consistent with the expansion of Acheulean tool technology, and a few others with other gene flows such as an expansion out of Eurasia and back into Africa subsequent to the most recent expansion out of Africa. Templeton rejected a hypothesis of recent African replacement with greater than 99% certainty (p < 10−17).[49]
Ancient DNA
Main article: Archaic Homo sapiens admixture with modern humansBy 2006, extraction of DNA directly from some archaic human samples was becoming possible. The earliest analyses were of neanderthal DNA, and indicated that the Neanderthal contribution to modern human genetic diversity was no more than 20%, with a most likely value of 0%.[50] By 2010, however, detailed DNA sequencing of the Neanderthal specimens from Europe indicated that the contribution was nonzero, with Neanderthals sharing 1-4% more genetic variants with living non-Africans than with living humans in sub-Saharan Africa, supporting regional continuity outside of Africa.[51][52] In late 2010, a recently discovered non-Neanderthal archaic human, the Denisova hominin from southern Siberia, was found to share 4-6% of its genome with living Melanesian humans and with no other living group, supporting lateral gene transfer between two regions outside of Africa.[53][54]
In August 2011, Denisovan and Neandertal archaic HLA types were found to represent more than half the HLA alleles of modern Eurasians.[55]
Recent African replacement
Main article: Recent African origin of modern humansThe primary competing scientific hypothesis is currently recent African origin of modern humans, which proposes that modern humans arose as a new species in Africa around 100-200,000 years ago, moving out of Africa around 50-60,000 years ago to replace existing human species such as Homo erectus and the Neanderthals without interbreeding.[56][57][58][59] This differs from the multiregional hypothesis in that the multiregional model predicts interbreeding with preexisting local human populations in any such migration.[59]
Dobzhansky paleovariabilism
A version of multiregional evolution (also a modification of polygenism that rejects independent origin of identical mutations) suggests that Homo sapiens evolved not through new mutations, but through selective pressure altering the statistical proportion between different alleles. Instead of the migrations suggested by Wolpoff, the Dobzhansky model suggests that all alleles needed to make a Homo sapiens already existed within the individual genetic variation of all Homo erectus populations by the time Homo erectus first migrated from Africa, albeit at a very low (but non-zero) frequency. The total absence of sapiens specimens from that time is explained by the fact that biological modernity is controlled by a multitude of genes (not by one single gene), and that the low frequency of modern alleles at that time means that not a single individual carried sufficient numbers of modern alleles to be considered a modern human[60].
See also
References
- ^ R.G. Bednarik (2011). "The Expulsion of Eve". Developments in Primatology: 25-55. doi:10.1007/978-1-4419-9353-3_2. http://www.springer.com/cda/content/document/cda_downloaddocument/9781441993526-c1.pdf?SGWID=0-0-45-1154940-p174101060. Retrieved 10 Nov 2011.
- ^ Blundell, Geoffrey (2006). Origins: the story of the emergence of humans and humanity in Africa. Double Storey. p. 69. ISBN 1770130403. http://books.google.ca/books?id=yzKeeReTRbwC&lpg=PA68&ots=29iHjRSWD-&dq=Origins%3A%20the%20story%20of%20the%20emergence%20of%20humans%20and%20humanity%20in%20Africa&pg=PA69#v=onepage&q&f=false.
- ^ Wolpoff, MH; JN Spuhler, FH Smith, J Radovcic, G Pope, DW Frayer, R Eckhardt, and G Clark (1988). "Modern human origins". Science 241 (4867): 772–774. Bibcode 1988Sci...241..772W. doi:10.1126/science.3136545. PMID 3136545. http://www.sciencemag.org/cgi/pdf_extract/241/4867/772.
- ^ a b Wolpoff, Milford, and Caspari, Rachel (1997). Race and Human Evolution. Simon & Schuster. pp. 28–29.
- ^ The people’s Peking man, popular science and human identity in twentieth-century China, Sigrid, Schmalzer, 2008 p. 252 ISBN 0226738604
- ^ Apes, Giants, and Man, Weidenreich, 1946 ISBN 0226881474
- ^ Wolpoff, MH; Hawks J, Caspari R (2000). "Multiregional, not multiple origins". Am J Phys Anthropol 112 (1): 129–36. doi:10.1002/(SICI)1096-8644(200005)112:1<129::AID-AJPA11>3.0.CO;2-K. PMID 10766948. http://www3.interscience.wiley.com/journal/71008905/abstract.
- ^ a b c Wolpoff, MH; JN Spuhler, FH Smith, J Radovcic, G Pope, DW Frayer, R Eckhardt, and G Clark (1988). "Modern Human Origins". Science 241 (4867): 772–4. http://dx.doi.org/10.1126%2Fscience.3136545
- ^ a b Wolpoff, Milford, and Caspari, Rachel (1997). Race and Human Evolution. Simon & Schuster. pp. 25–26.
- ^ Wolpoff, Milford H; John Hawks, David W Frayer, Keith Hunley (2001). "Modern Human Ancestry at the Peripheries: A Test of the Replacement Theory". Science (AAAS) 291 (5502): 293–297. Bibcode 2001Sci...291..293W. doi:10.1126/science.291.5502.293. PMID 11209077. http://www.sciencemag.org/cgi/content/abstract/291/5502/293.
- ^ Analysis of the full brain case shape confirms the idea that dispersal from a single population could not explain the early modern human variability, but does not confirm ties to regional archaic humans. Gunz, P., et. al. (2009-04-14). "Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario". Proceedings of the National Academy of Sciences of the United States of America 106 (15): 6094–6098. Bibcode 2009PNAS..106.6094G. doi:10.1073/pnas.0808160106. PMC 2669363. PMID 19307568. http://www.pnas.org/content/106/15/6094.abstract.
- ^ Shang et al.; Tong, H; Zhang, S; Chen, F; Trinkaus, E (1999). "An early modern human from Tianyuan Cave, Zhoukoudian, China" (Free full text). Proceedings of the National Academy of Sciences of the United States of America 104 (16): 6573–8. Bibcode 2007PNAS..104.6573S. doi:10.1073/pnas.0702169104. ISSN 0027-8424. PMC 1871827. PMID 17416672. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17416672.
- ^ Phil McKenna, "Chinese challenge to 'out of Africa' theory", New Scientist, Nov 2009
- ^ Wolpoff, Milford; Bruce Mannheim, Alan Mann, John Hawks, Rachel Caspari, Karen R. Rosenberg, David W. Frayer, George W. Gill and Geoffrey Clark (2004). "Why not the Neandertals?". World Archaeology 36 (4): 527. doi:10.1080/0043824042000303700.
- ^ Harvati, Katerina; Stephen R Frost and Kieran P McNulty (2004). "Neanderthal taxonomy reconsidered: Implications of 3D primate models of intra- and interspecific differences". PNAS 101 (5): 1147–1152. Bibcode 2004PNAS..101.1147H. doi:10.1073/pnas.0308085100. PMC 337021. PMID 14745010. http://www.pnas.org/content/101/5/1147.full.
- ^ Pearson, Osbjorn M (2004). "Has the Combination of Genetic and Fossil Evidence Solved the Riddle of Modern Human Origins?". Evolutionary Anthropology 13 (4): 145–159. doi:10.1002/evan.20017.
- ^ Duarte C, 2. Maurício J, Pettitt P, Souto P, Trinkaus E, van der Plicht H, Zilhão J (1999). "The early Upper Paleolithic human skeleton from the Abrigo do Lagar Velho (Portugal) and modern human emergence in Iberia". Proc Natl Acad Sci USA 96 (13): 7604–7609. Bibcode 1999PNAS...96.7604D. doi:10.1073/pnas.96.13.7604. PMC 22133. PMID 10377462. http://www.pnas.org/content/96/13/7604.abstract?ijkey=9335ab52731624a02b5f7f426c4a8c2147934993&keytype2=tf_ipsecsha.
- ^ Trinkaus, E; Moldovan, O; Milota, S; Bîlgăr, A; Sarcina, L; Athreya, S; Bailey, Se; Rodrigo, R; Mircea, G; Higham, T; Ramsey, Cb; Van, Der, Plicht, J (Sep 2003). "An early modern human from the Peştera cu Oase, Romania" (free full text). Proceedings of the National Academy of Sciences of the United States of America 100 (20): 11231–11236. Bibcode 2003PNAS..10011231T. doi:10.1073/pnas.2035108100. ISSN 0027-8424. PMC 208740. PMID 14504393. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=14504393. "When multiple measurements are undertaken, the mean result can be determined through averaging the activity ratios. For Oase 1, this provides a weighted average activity ratio of 〈14a〉 = 1.29 ± 0.15%, resulting in a combined OxA-GrA 14C age of 34,950, +990, and –890 B.P."
- ^ Tattersall, Ian, and Schwartz, Jeffrey H. (1999). "Hominids and hybrids: The place of Neanderthals in human evolution". Proceedings of the National Academy of Sciences of the United States of America 96 (13): 7117–7119. Bibcode 1999PNAS...96.7117T. doi:10.1073/pnas.96.13.7117. PMC 33580. PMID 10377375. http://www.pnas.org/cgi/content/full/96/13/7117.
- ^ Trinkaus, E (May 2007). "European early modern humans and the fate of the Neandertals" (Free full text). Proceedings of the National Academy of Sciences of the United States of America 104 (18): 7367–72. Bibcode 2007PNAS..104.7367T. doi:10.1073/pnas.0702214104. ISSN 0027-8424. PMC 1863481. PMID 17452632. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17452632.
- ^ http://www.sciencedaily.com/releases/2007/04/070423185434.htm The Emerging Fate Of The Neandertals
- ^ Cann, Rebecca L., Stoneking, Mark, and Wilson, Allan C. (1987-01-01). "Mitochondrial DNA and human evolution". Nature 325 (6099): 31–36. Bibcode 1987Natur.325...31C. doi:10.1038/325031a0. PMID 3025745. http://artsci.wustl.edu/~landc/html/cann/.
- ^ Hodgson, JA; Disotell TR (2008). "No evidence of a Neanderthal contribution to modern human diversity.". Genome Biology (BioMed Central) 9 (2): 206. doi:10.1186/gb-2008-9-2-206. PMC 2374707. PMID 18304371. http://genomebiology.com/2008/9/2/206.
- ^ Krause, Johannes, et al (2010-03-24). "The complete mitochondrial DNA genome of an unknown hominin from southern Siberia". Nature 464 (7290): 894–897. Bibcode 2010Natur.464..894K. doi:10.1038/nature08976. PMID 20336068.
- ^ Wolpoff, Milford, and Caspari, Rachel (1997). Race and Human Evolution. Simon & Schuster. pp. 30.
- ^ Relethford, JH (2008-03-05). "Genetic evidence and the modern human origins debate". Heredity (Macmillan) 100 (6): 555–63. doi:10.1038/hdy.2008.14. PMID 18322457.
- ^ "Selection, nuclear genetic variation, and mtDNA". john hawks weblog. http://johnhawks.net/weblog/reviews/neandertals/neandertal_dna/weaver_roseman_2005.html. Retrieved 2011-01-05.
- ^ Hammer, M. F., et. al." (1998). "Out of Africa and Back Again: Nested Cladistic Analysis of Human Y Chromosome Variation". Molecular Biology and Evolution 15 (4): 427–441. PMID 9549093. http://mbe.oxfordjournals.org/content/15/4/427.short.
- ^ Hammer, Mf; Garrigan, D; Wood, E; Wilder, Ja; Mobasher, Z; Bigham, A; Krenz, Jg; Nachman, Mw (Aug 2004). "Heterogeneous patterns of variation among multiple human x-linked Loci: the possible role of diversity-reducing selection in non-africans" (Free full text). Genetics 167 (4): 1841–53. doi:10.1534/genetics.103.025361. ISSN 0016-6731. PMC 1470985. PMID 15342522. http://www.genetics.org/cgi/content/abstract/167/4/1841?ijkey=cb14a3724516d1a584feb8454d2c49cd72e003ee&keytype2=tf_ipsecsha. Additional discussion of these results is available in a video of a presentation given by Hammer at http://www.youtube.com/watch?v=Ff0jwWaPlnU (video) from about 40 to 50 minutes into the video.
- ^ The CMP-N-acetylneuraminic acid hydroxylase CMAH pseudogene shows 2.9 Mya coalescence time. Hayakawa, T; Aki, I; Varki, A; Satta, Y; Takahata, N (Feb 2006). "Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution". Genetics 172 (2): 1139–46. doi:10.1534/genetics.105.046995. ISSN 0016-6731. PMC 1456212. PMID 16272417. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1456212.
- ^ The ALMS1 site suggests ancient and complex evolutionary history with a coalescence time of about 2 Mya. Scheinfeldt, Lb; Biswas, S; Madeoy, J; Connelly, Cf; Schadt, Ee; Akey, Jm (Jun 2009). "Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history". Molecular biology and evolution 26 (6): 1357–67. doi:10.1093/molbev/msp045. ISSN 0737-4038. PMC 2734137. PMID 19279085. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2734137.
- ^ The PDHA1 (pyruvate dehydrogenase) locus on the X chromosome has an estimated coalescence time of 1.86 Mya, inconsistent with a recent species origin, although the worldwide lineage pattern is unlike other autosomal sites and may be consistent with recent dispersal from Africa. Rosalind M. Harding (March 16, 1999). "More on the X files". Proceedings of the National Academy of Sciences 96 (6): 2582–2584. Bibcode 1999PNAS...96.2582H. doi:10.1073/pnas.96.6.2582. http://www.pnas.org/cgi/content/full/96/6/2582.
- ^ A second group finds the same ancient origin for PDHA1, but finds no evidence of a recent expansion, consistent with other autosomal and X chromosome sites and contrary to mitochondrial data. Harris, E. E.; Jody Hey (1999). "X chromosome evidence for ancient human histories". Proceedings of the National Academy of Sciences 96 (6): 3320–4. Bibcode 1999PNAS...96.3320H. doi:10.1073/pnas.96.6.3320. PMC 15940. PMID 10077682. http://www.pnas.org/content/96/6/3320.full.
- ^ The ASAH1 gene has two recently differentiated lineages with a coalescence time 2.4±.4 Mya not explainable by balancing selection. The V lineage shows evidence of recent positive selection. The lineage pattern may be the result of hybridization during a recent range expansion from Africa with the V lineage tracing to archaic humans from outside Africa, though it is also consistent with a mixture of two long isolated groups within Africa; it is not consistent with a recent origination of a modern human species that replaced archaic forms without interbreeding. Kim, Hl; Satta, Y (Mar 2008). "Population genetic analysis of the N-acylsphingosine amidohydrolase gene associated with mental activity in humans" (Free full text). Genetics 178 (3): 1505–15. doi:10.1534/genetics.107.083691. ISSN 0016-6731. PMC 2278054. PMID 18245333. http://www.genetics.org/cgi/pmidlookup?view=long&pmid=18245333. ""It is speculated that, when modern humans dispersed from Africa, admixture of the distinct V and M lineages occurred and the V lineage has since spread in the entire population by possible positive selection.""
- ^ Daniel Garrigan, Zahra Mobasher, Sarah B. Kingan, Jason A. Wilder and Michael F. Hammer (Aug 2005). "Deep Haplotype Divergence and Long-Range Linkage Disequilibrium at Xp21.1 Provide Evidence That Humans Descend From a Structured Ancestral Population". Genetics 170 (4): 1849–1856. doi:10.1534/genetics.105.041095. PMC 1449746. PMID 15937130. http://www.genetics.org/cgi/content/abstract/170/4/1849.
- ^ NAT2 SNPs lineages cluster in sub-Saharan Africa, Europe, and East Asia, with genetic distances scaling with geographic distances. Sabbagh, A; Langaney, A; Darlu, P; Gérard, N; Krishnamoorthy, R; Poloni, Es (Feb 2008). "Worldwide distribution of NAT2 diversity: implications for NAT2 evolutionary history" (Free full text). BMC genetics 9: 21. doi:10.1186/1471-2156-9-21. PMC 2292740. PMID 18304320. http://www.biomedcentral.com/1471-2156/9/21. Also see map; may resize browser window.
- ^ The NAT1 lineage tree is rooted in Eurasia with a coalescence time of 2.0 Mya that cannot be explained by balancing selection and with the NAT1*11A haplotype absent from subsaharan Africa. Patin, E; Barreiro, Lb; Sabeti, Pc; Austerlitz, F; Luca, F; Sajantila, A; Behar, Dm; Semino, O; Sakuntabhai, A; Guiso, N; Gicquel, B; Mcelreavey, K; Harding, Rm; Heyer, E; Quintana-Murci, L (Mar 2006). "Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes". American journal of human genetics 78 (3): 423–36. doi:10.1086/500614. ISSN 0002-9297. PMC 1380286. PMID 16416399. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1380286.
- ^ johnhawks.net "Variation in NAT1 and NAT2". john hawks weblog. http://johnhawks.net/weblog/reviews/genetics/diet/nat1_nat2_patin_2006_selection.html johnhawks.net. Retrieved 2011-01.04.
- ^ Garrigan, D; Mobasher, Z; Severson, T; Wilder, Ja; Hammer, Mf (Feb 2005). "Evidence for archaic Asian ancestry on the human X chromosome" (Free full text). Molecular biology and evolution 22 (2): 189–92. doi:10.1093/molbev/msi013. ISSN 0737-4038. PMID 15483323. http://mbe.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=15483323.
- ^ Cox, Mp; Mendez, Fl; Karafet, Tm; Pilkington, Mm; Kingan, Sb; Destro-Bisol, G; Strassmann, Bi; Hammer, Mf (Jan 2008). "Testing for archaic hominin admixture on the X chromosome: model likelihoods for the modern human RRM2P4 region from summaries of genealogical topology under the structured coalescent" (Free full text). Genetics 178 (1): 427–37. doi:10.1534/genetics.107.080432. ISSN 0016-6731. PMC 2206091. PMID 18202385. http://www.genetics.org/cgi/pmidlookup?view=long&pmid=18202385.
- ^ J. Hardy, A. Pittman, A. Myers, K. Gwinn-Hardy, H.C. Fung, R. de Silva, M. Hutton and J. Duckworth (2005). "Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens". Biochemical Society Transactions 33, part 4: 582–585. http://www.biochemsoctrans.org/bst/033/0582/0330582.pdf.
- ^ Zody, Mc; Jiang, Z; Fung, Hc; Antonacci, F; Hillier, Lw; Cardone, Mf; Graves, Ta; Kidd, Jm; Cheng, Z; Abouelleil, A; Chen, L; Wallis, J; Glasscock, J; Wilson, Rk; Reily, Ad; Duckworth, J; Ventura, M; Hardy, J; Warren, Wc; Eichler, Ee (Aug 2008). "Evolutionary toggling of the MAPT 17q21.31 inversion region". Nature genetics 40 (9): 1076–83. doi:10.1038/ng.193. ISSN 1061-4036. PMC 2684794. PMID 19165922. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2684794.
- ^ Almos, Pz; Horváth, S; Czibula, A; Raskó, I; Sipos, B; Bihari, P; Béres, J; Juhász, A; Janka, Z; Kálmán, J (Nov 2008). "H1 tau haplotype-related genomic variation at 17q21.3 as an Asian heritage of the European Gypsy population". Heredity 101 (5): 416–9. doi:10.1038/hdy.2008.70. ISSN 0018-067X. PMID 18648385.
- ^ Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et. al. (2005-01-16). "A common inversion under selection in Europeans". Nature genetics 37 (2): 129–137. doi:10.1038/ng1508. PMID 15654335. http://www.genetics.org/cgi/content/abstract/170/4/1849.
- ^ Evans, Pd; Mekel-Bobrov, N; Vallender, Ej; Hudson, Rr; Lahn, Bt (Nov 2006). "Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage" (Free full text). Proceedings of the National Academy of Sciences of the United States of America 103 (48): 18178–83. Bibcode 2006PNAS..10318178E. doi:10.1073/pnas.0606966103. ISSN 0027-8424. PMC 1635020. PMID 17090677. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17090677.
- ^ Trinkaus, E (May 2007). "European early modern humans and the fate of the Neandertals" (Free full text). Proceedings of the National Academy of Sciences of the United States of America 104 (18): 7367–72. Bibcode 2007PNAS..104.7367T. doi:10.1073/pnas.0702214104. ISSN 0027-8424. PMC 1863481. PMID 17452632. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17452632.
- ^ Evans, Pd; Gilbert, Sl; Mekel-Bobrov, N; Vallender, Ej; Anderson, Jr; Vaez-Azizi, Lm; Tishkoff, Sa; Hudson, Rr; Lahn, Bt (Sep 2005). "Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans". Science 309 (5741): 1717–20. Bibcode 2005Sci...309.1717E. doi:10.1126/science.1113722. ISSN 0036-8075. PMID 16151009.
- ^ "Introgression and microcephalin FAQ". john hawks weblog. http://johnhawks.net/weblog/reviews/neandertals/neandertal_dna/introgression_faq_2006.html. Retrieved 2011-01-05.
- ^ Templeton, Alan R. (2005). "Haplotype Trees and Modern Human Origins". Yearbook of Physical Anthropology 48 (S41): 33–59. doi:10.1002/ajpa.20351. http://esa.ipb.pt/pdf/28.pdf.
- ^ Noonan, James P., et. al. (2006-11-17). "Sequencing and Analysis of Neanderthal Genomic DNA". Science 314 (5802): 1113–1118. Bibcode 2006Sci...314.1113N. doi:10.1126/science.1131412. PMC 2583069. PMID 17110569. http://www.sciencemag.org/content/328/5979/710.
- ^ Green, Richard E., et. al. (2010-05-07). "A Draft Sequence of the Neandertal Genome". Science 328 (5979): 710–722. Bibcode 2010Sci...328..710G. doi:10.1126/science.1188021. PMID 20448178. http://www.sciencemag.org/content/328/5979/710.
- ^ "NEANDERTALS LIVE!". john hawks weblog. http://johnhawks.net/weblog/reviews/neandertals/neandertal_dna/neandertals-live-genome-sequencing-2010.html. Retrieved 2010-12-31.
- ^ Reich, David, et al (2010-12-23). "Genetic history of an archaic hominin group from Denisova Cave in Siberia". Nature 468 (7327): 1053–1060. Bibcode 2010Natur.468.1053R. doi:10.1038/nature09710. PMID 21179161. http://www.nature.com/nature/journal/v468/n7327/full/nature09710.html.
- ^ "The Denisova genome FAQ". john hawks weblog. http://johnhawks.net/weblog/reviews/neandertals/neandertal_dna/denisova-nuclear-genome-reich-2010.html. Retrieved 2010-12-31.
- ^ Laurent Abi-Rached, et. al. (2011-08-25). "The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans". Science 334 (6052). doi:10.1126/science.1209202. Archived from the original on Aug 2011. http://digitalcommons.unl.edu/cgi/viewcontent.cgi?article=1122&context=publichealthresources.
- ^ Hua Liu, et al. (2006). "A Geographically Explicit Genetic Model of Worldwide Human-Settlement History". American Journal of Human Genetics 79 (2): 230–237. doi:10.1086/505436. PMC 1559480. PMID 16826514. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1559480.
- ^ Weaver, Timothy D; Charles C. Roseman (2008). "New developments in the genetic evidence for modern human origins". Evolutionary Anthropology: Issues, News, and Reviews (Wiley-Liss) 17 (1): 69–80. doi:10.1002/evan.20161. http://www3.interscience.wiley.com/journal/117921411/abstract.
- ^ Fagundes, NJ; Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L. (2007). "Statistical evaluation of alternative models of human evolution". Proc Natl Acad Sci USA 104 (45): 17614–9. Bibcode 2007PNAS..10417614F. doi:10.1073/pnas.0708280104. PMC 2077041. PMID 17978179. http://www.pnas.org/content/104/45/17614.long.
- ^ a b Wolpoff, Milford, and Caspari, Rachel (1997). Race and Human Evolution. Simon & Schuster. p. 42.
- ^ Book recension of Carleton Coon´s book "The origin of races": Recension by Theodosius Dobzhansky
External links
- From www.conrante.com: Templeton's lattice diagram showing major gene flows graphically.
- From darwin.eeb.uconn.edu: notes on drift and migration with equations for calculating the effects on allele frequencies of different populations.
- Human Evolution. (2011). In Encyclopædia Britannica. Retrieved from http://www.britannica.com/EBchecked/topic/275670/human-evolution
Part of the series on Human evolution Humans and Proto-humansHomo: H. gautengensis · H. habilis · H. rudolfensis · H. georgicus · H. ergaster · H. erectus (H. e. erectus · H. e. lantianensis · H. e. palaeojavanicus · H. e. pekinensis · H. e. nankinensis · H. e. wushanensis · H. e. yuanmouensis · H. e. soloensis) · H. cepranensis · H. antecessor · H. heidelbergensis · Denisova hominin · H. neanderthalensis · H. rhodesiensis · H. floresiensis · Archaic Homo sapiens · Anatomically modern humans (H. s. idaltu · H. s. sapiens)Topics: Timeline of human evolution · List of human evolution fossils · Human evolutionary genetics
Models: Recent African origin · Multiregional originCategories:
Wikimedia Foundation. 2010.