- DNA clamp
-
Top and side views of a homotrimer of the human PCNA sliding clamp (rainbow colored, N-terminus = blue, C-terminus = red) with double stranded DNA modeled through the central pore (magenta).[1]
A DNA clamp, also known as a sliding clamp, is a protein fold that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because the rate-limiting step in the DNA synthesis reaction is the association of the polymerase with the DNA template,[citation needed] the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.
Contents
Structure
The DNA clamp fold is an α+β protein that assembles into a multimeric structure that completely encircles the DNA double helix as the polymerase adds nucleotides to the growing strand.[2] The DNA clamp assembles on the DNA at the replication fork and "slides" along the DNA with the advancing polymerase, aided by a layer of water molecules in the central pore of the clamp between the DNA and the protein surface. Because of the toroidal shape of the assembled multimer, the clamp cannot dissociate from the template strand without also dissociating into monomers.
The DNA clamp fold is found in bacteria, archaea, eukaryotes and some viruses. In bacteria, the sliding clamp is a homodimer composed of two identical beta subunits of DNA polymerase III and hence is referred to as the beta clamp. In archaea[3] and eukaryotes, it is a trimer composed of three molecules of PCNA. The T4 bacteriophage also uses a sliding clamp, called gp45 that is a trimer similar in structure to PCNA but lacks sequence homology to either PCNA or the bacterial beta clamp.[2]
Kingdom Sliding clamp protein Aggregation state Associated polymerase Bacteria beta subunit of pol III dimer DNA polymerase III Archaea archaeal PCNA trimer pol ε Eukaryote PCNA trimer DNA polymerase delta Virus gp43 / gp45 timer RB69 Pol / T4 Pol Bacterial
DNA polymerase III subunit beta 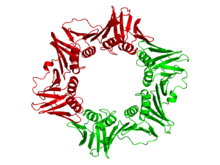
Crystallographic structure of the dimeric DNA polymerase beta subunit from E. coli.[4] Identifiers Organism Symbol dnaN Entrez 948218 PDB 1MMI RefSeq (Prot) NP_418156 UniProt P0A988 Other data EC number 2.7.7.7 Chromosome MG1655: 3.88 - 3.88 Mb The beta clamp is a specific DNA clamp and a subunit of the DNA polymerase III holoenzyme found in bacteria. Two beta subunits are assembled around the DNA by the gamma subunit and ATP hydrolysis; this assembly is called the pre-initiation complex. After assembly around the DNA, the beta subunits' affinity for the gamma subunit is replaced by an affinity for the alpha and epsilon subunits, which together create the complete holoenzyme.[5][6][7] DNA polymerase III is the primary enzyme complex involved in prokaryotic DNA replication.
The gamma complex of DNA polymerase III, composed of γδδ'χψ subunits, catalyzes ATP to chaperone two beta subunits to bind to DNA. Once bound to DNA, the beta subunits can freely slide along double stranded DNA. The beta subunits in turn bind the αε polymerase complex. The α subunit possesses DNA polymerase activity and the ε subunit is a 3’-5’ exonuclease.[7]
The beta chain of bacterial DNA polymerase III is composed of three topologically non-equivalent domains (N-terminal, central, and C-terminal). Two beta chain molecules are tightly associated to form a closed ring encircling duplex DNA.
DNA polymerase III, beta chain Identifiers Symbol DNA_polIII_beta Pfam PF00712 InterPro IPR001001 SMART SM00480 SCOP 2pol Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary DNA polymerase III, beta chain,
N-terminalIdentifiers Symbol DNA_pol3_beta Pfam PF00712 InterPro IPR022634 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary DNA polymerase III, beta chain,
centralIdentifiers Symbol DNA_pol3_beta_2 Pfam PF02767 InterPro IPR022637 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary DNA polymerase III, beta chain,
C-terminalIdentifiers Symbol DNA_pol3_beta_3 Pfam PF02768 InterPro IPR022635 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Eukaryote
proliferating cell nuclear antigen 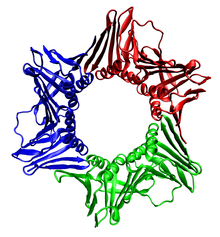
The assembled human DNA clamp, a trimer of the human protein PCNA.[8] Identifiers Symbol PCNA Entrez 5111 HUGO 8729 OMIM 176740 PDB 1axc RefSeq NM_002592 UniProt P12004 Other data EC number 2.7.7.7 Locus Chr. 20 pter-p12 The sliding clamp in eukaryotes is assembled from a specific subunit of DNA polymerase delta called the proliferating cell nuclear antigen (PCNA). The N-terminal and C-terminal domains of PCNA are topologically identical. Three PCNA molecules are tightly associated to form a closed ring encircling duplex DNA.
The sequence of PCNA is well conserved between plants and animals, indicating a strong selective pressure for structure conservation, and suggesting that this type of DNA replication mechanism is conserved throughout eukaryotes.[9] Homologues of PCNA have also been identified in the archaea (Euryarchaeota and Crenarchaeota) and in Paramecium bursaria Chlorella virus 1 (PBCV-1) and in nuclear polyhedrosis viruses.
Proliferating cell nuclear antigen, N-terminal domain Identifiers Symbol PCNA_N Pfam PF00705 InterPro IPR000730 PROSITE PDOC00265 SCOP 1plq Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Proliferating cell nuclear antigen, C-terminal domain Identifiers Symbol PCNA_C Pfam PF02747 InterPro IPR000730 PROSITE PDOC00265 SCOP 1plq Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Viral
DNA polymerase accessory protein 45 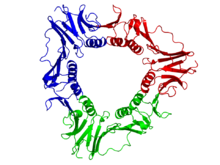
Crystallographic structure of the trimeric gp45 sliding clamp from bacteriophage T4.[10] Identifiers Organism Symbol gp45 Entrez 1258821 PDB 1CZD RefSeq (Prot) NP_049666 UniProt P04525 Other data EC number 2.7.7.7 Chromosome 1: 0.03 - 0.03 Mb The viral gp45 sliding clamp subunit protein contains two domains. Each domain consists of two alpha helices and two beta sheets – the fold is duplicated and has internal pseudo two-fold symmetry.[11] Three gp45 molecules are tightly associated to form a closed ring encircling duplex DNA.
Gp45 sliding clamp, N-terminal Identifiers Symbol DNA_pol_proc_fac Pfam PF02916 InterPro IPR004190 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Gp45 sliding clamp, C-terminal Identifiers Symbol Gp45_slide_clamp_C Pfam PF09116 InterPro IPR015200 Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary Assembly
Sliding clamps are loaded onto their associated DNA template strands by specialized proteins known as "sliding clamp loaders", which also disassemble the clamps after replication has completed. The binding sites for these initiator proteins overlap with the binding sites for the DNA polymerase, so the clamp cannot simultaneously associate with a clamp loader and with a polymerase. Thus the clamp will not be actively disassembled while the polymerase remains bound. Although DNA clamps play a less significant role in associating with other DNA-interacting proteins, such as nucleosome assembly factors, Okazaki fragment ligases, and DNA repair proteins, all of these proteins also share a binding site on the DNA clamp that overlaps with the clamp loader site, ensuring that the clamp will not be removed while any enzyme is still working on the DNA. The activity of the clamp loader requires ATP hydrolysis to "close" the clamp around the DNA.
References
- ^ PDB 1W60; Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, Fischer PM, Walkinshaw MD (February 2005). "Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design". Proc. Natl. Acad. Sci. U.S.A. 102 (6): 1871–6. doi:10.1073/pnas.0406540102. PMC 548533. PMID 15681588. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=548533.
- ^ a b Bruck I, O'Donnell M (2001). "The ring-type polymerase sliding clamp family". Genome Biol. 2 (1): REVIEWS3001. doi:10.1186/gb-2001-2-1-reviews3001. PMC 150441. PMID 11178284. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=150441.
- ^ Matsumiya S, Ishino Y, Morikawa K (January 2001). "Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus". Protein Sci. 10 (1): 17–23. doi:10.1110/ps.36401. PMC 2249843. PMID 11266590. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2249843.
- ^ PDB 1MMI; Oakley AJ, Prosselkov P, Wijffels G, Beck JL, Wilce MC, Dixon NE (July 2003). "Flexibility revealed by the 1.85 Å crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III". Acta Crystallogr. D Biol. Crystallogr. 59 (Pt 7): 1192–9. doi:10.1107/S0907444903009958. PMID 12832762.
- ^ Lewin, Benjamin (1997). Genes VI. Oxford [Oxfordshire]: Oxford University Press. pp. 484–7. ISBN 0-19-857779-6.
- ^ Lehninger, Albert L (1975). Biochemistry: The Molecular Basis of Cell Structure and Function. New York: Worth Publishers. pp. 894. ISBN 0-87901-047-9.
- ^ a b Stukenberg PT, Studwell-Vaughan PS, O'Donnell M (June 1991). "Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme". J. Biol. Chem. 266 (17): 11328–34. PMID 2040637. http://www.jbc.org/content/266/17/11328.abstract.
- ^ PDB 1AXC; Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J (October 1996). "Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA". Cell 87 (2): 297–306. doi:10.1016/S0092-8674(00)81347-1. PMID 8861913.
- ^ Suzuka I, Hata S, Matsuoka M, Kosugi S, Hashimoto J (January 1991). "Highly conserved structure of proliferating cell nuclear antigen (DNA polymerase delta auxiliary protein) gene in plants". Eur. J. Biochem. 195 (2): 571–5. doi:10.1111/j.1432-1033.1991.tb15739.x. PMID 1671766.
- ^ PDB 1CZD; Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J (March 2000). "Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage". J. Mol. Biol. 296 (5): 1215–23. doi:10.1006/jmbi.1999.3511. PMID 10698628.
- ^ Steitz TA, Shamoo Y (1999). "Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex". Cell 99 (2): 155–166. doi:10.1016/S0092-8674(00)81647-5. PMID 10535734.
Further reading
- Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R (2004). Molecular Biology of the Gene. San Francisco: Pearson/Benjamin Cummings. ISBN 0-8053-4635-X.
External links
General All-α folds: All-β folds: α/β folds: α+β folds: Irregular folds: ←Secondary structureCategories:- Genes on chromosome 20
- Biotechnology
- Protein folds
- DNA replication
Wikimedia Foundation. 2010.


