- Propionyl-CoA carboxylase
-
"PCCA" redirects here. For other uses, see PCCA (disambiguation).
Propionyl-CoA carboxylase Identifiers EC number 6.4.1.3 CAS number 9023-94-3 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles Methylmalonyl-CoA decarboxylase Identifiers EC number 4.1.1.41 CAS number 37289-44-4 Databases IntEnz IntEnz view BRENDA BRENDA entry ExPASy NiceZyme view KEGG KEGG entry MetaCyc metabolic pathway PRIAM profile PDB structures RCSB PDB PDBe PDBsum Gene Ontology AmiGO / EGO Search PMC articles PubMed articles Propionyl-CoA carboxylase catalyses the carboxylation reaction of propionyl CoA in the mitochondrial matrix. The enzyme is biotin dependent. The product of the reaction is (S)-methylmalonyl CoA. Propionyl CoA is the end product of metabolism of odd-chain fatty acids, and is also a metabolite of most methyl-branched fatty acids. It is also the main metabolite of valine, and together with acetyl-CoA, is a metabolite of isoleucine, as well as a methionine metabolite. Propionyl-CoA is thus of great importance as a glucose precursor. (S)-Methylmalonyl-CoA is not directly utilizable by animals; it is acted on by a racemase to give (R)-methylmalonyl-CoA. The latter is converted by methylmalonyl-CoA mutase (one of a very few Vitamin B12 dependent enzymes) to give succinyl-CoA. The latter is converted to oxaloacetate and then malate in the Krebs cycle. Export of malate into the cytosol leads to formation of oxaloacetate, phosphoenol pyruvate, and other gluconeogenic intermediates.
- ATP + propanoyl-CoA + HCO3- <=> ADP + phosphate + (S)-methylmalonyl-CoA
It has been classified both as a ligase[1] and a lyase.[2]
Contents
Enzyme Structure
Propionyl-CoA Carboxylase (PCC) is a 750 kDa alpha(6)-beta(6)-dodecamer. (Only approximately 540 kDa is native enzyme. [3] ) The alpha subunits are arranged as monomers, decorating the central beta-6 hexameric core. Said core is oriented as a short cylinder with a hole along its axis.
The alpha subunit of PCC contains the biotin carboxylase (BC) and biotin carboxyl carrier protein (BCCP) domains. A domain known as the BT domain is also located on the alpha subunit and is essential for interactions with the beta subunit. The 8-stranded anti-parallel beta barrel fold of this domain is particularly interesting. The beta subunit contains the carboxyltransferase (CT) activity.[4]
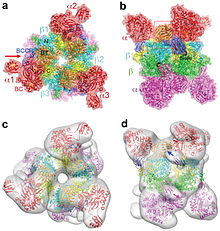 Figure 1.(a). Schematic drawing of the structure of the RpPCCα-RdPCCβ chimera, viewed down the three-fold symmetry axis. Domains in the α and β subunits in the top half of the structure are given different colors, and those in the first α and β subunits are labeled. The α and β subunits in the bottom half are colored in magenta and green, respectively. The red arrow indicates the viewing direction of panel b. (b). Structure of the RpPCCα-RdPCCβ chimera, viewed down the two-fold symmetry axis. The red rectangle indicates the region shown in detail in Fig. 2a. (c). Cryo-EM reconstruction of HsPCC at 15 Å resolution, viewed in the same orientation as panel a. The atomic model of the chimera was fit into the cryo-EM envelope. (d). The cryo-EM reconstruction viewed in the same orientation as panel b. The arrows indicate a change in the BCCP position that is needed to fit the cryo-EM map. All the structure figures were produced with PyMOL (www.pymol.org), and the cryo-EM figures were produced with Chimera.[5] This provides clear evidence of crucial dimeric interaction between alpha and beta subunits.
Figure 1.(a). Schematic drawing of the structure of the RpPCCα-RdPCCβ chimera, viewed down the three-fold symmetry axis. Domains in the α and β subunits in the top half of the structure are given different colors, and those in the first α and β subunits are labeled. The α and β subunits in the bottom half are colored in magenta and green, respectively. The red arrow indicates the viewing direction of panel b. (b). Structure of the RpPCCα-RdPCCβ chimera, viewed down the two-fold symmetry axis. The red rectangle indicates the region shown in detail in Fig. 2a. (c). Cryo-EM reconstruction of HsPCC at 15 Å resolution, viewed in the same orientation as panel a. The atomic model of the chimera was fit into the cryo-EM envelope. (d). The cryo-EM reconstruction viewed in the same orientation as panel b. The arrows indicate a change in the BCCP position that is needed to fit the cryo-EM map. All the structure figures were produced with PyMOL (www.pymol.org), and the cryo-EM figures were produced with Chimera.[5] This provides clear evidence of crucial dimeric interaction between alpha and beta subunits.
The BC and CT sites are approximately 55 Å apart, indicative of the entire BCCP domain translocating during catalysis of the carboxylation of propionyl-CoA. [5] This provides clear evidence of crucial dimeric interaction between alpha and beta subunits.
 Figure 2.(a). Schematic drawing of the relative positioning of the BC and CT active sites in the holoenzyme. One α subunit and a β2 dimer (β1 from one layer and β4 from the other layer) are shown, and the viewing direction is the same as Fig. 1b. The two active sites are indicated with the stars, separated by 55 Å distance. The bound positions of ADP in complex with E. coli BC 18 and that of CoA in complex with the 12S subunit of transcarboxylase 21 are also shown. (b). Detailed interactions between BCCP-biotin and the C domain of a β subunit. Hydrogen-bonding interactions are indicated with the dashed lines in red. The N1′ atom of biotin is labeled as 1′, hydrogen-bonded to the main-chain carbonyl of Phe397. (c). Molecular surface of the CT active site, showing a deep canyon where both substrates are bound. (d). Schematic drawing of the CT active site.[5]
Figure 2.(a). Schematic drawing of the relative positioning of the BC and CT active sites in the holoenzyme. One α subunit and a β2 dimer (β1 from one layer and β4 from the other layer) are shown, and the viewing direction is the same as Fig. 1b. The two active sites are indicated with the stars, separated by 55 Å distance. The bound positions of ADP in complex with E. coli BC 18 and that of CoA in complex with the 12S subunit of transcarboxylase 21 are also shown. (b). Detailed interactions between BCCP-biotin and the C domain of a β subunit. Hydrogen-bonding interactions are indicated with the dashed lines in red. The N1′ atom of biotin is labeled as 1′, hydrogen-bonded to the main-chain carbonyl of Phe397. (c). Molecular surface of the CT active site, showing a deep canyon where both substrates are bound. (d). Schematic drawing of the CT active site.[5]
The biotin-binding pocket of PCC is hydrophobic and highly conserved. Biotin and propionyl-CoA bind perpendicular to each other in the oxyanion hole containing active site. The native enzyme to biotin ratio has been determined to be one mole native enzyme to 4 moles biotin. [6] The N1 of biotin is thought to be the active site base.[7]
Site-directed mutagenesis at D422 shows a change in the substrate specificity of the propionyl-CoA binding site, thus indicating this residue’s importance in PCC’s catalytic activity. [8] In 1979, inhibition by phenylglyoxal determined that a phosphate group from either propionyl-CoA or ATP reacts with an essential arginine residue in the active site during catalysis.[9] Later (2004), it was suggested that Arginine-338 serves to orient the carboxyphosphate intermediate for optimal carboxylation of biotin. [10]
The KM values for ATP, propionyl-CoA, and bicarbonate has been determined to be 0.08 mM, 0.29 mM, and 3.0 mM respectively. The isoelectric point falls at pH 5.5. PCC’s structural integrity is conserved over the temperature range of -50 to 37 degrees Celsius and the pH range of 6.2 to 8.8. Optimum pH was shown to be between 7.2 and 8.8 without biotin bound.[11] With biotin, optimum pH is 8.0-8.5.[12]
Enzyme Mechanism
The normal catalytic reaction mechanism involves a carbanion intermediate and does not proceed through a concerted process.[13] Figure 3 shows a probable pathway.
 Figure 3. Probable PCC Mechanism
Figure 3. Probable PCC Mechanism
The reaction has been shown to be slightly reversible at low propionyl-CoA flux.[14]
Isozymes
Humans express the following two propionyl-CoA carboxylase isozymes:
propionyl Coenzyme A carboxylase, alpha polypeptide Identifiers Symbol PCCA Entrez 5095 HUGO 8653 OMIM 232000 RefSeq NM_000282 UniProt P05165 Other data EC number 6.4.1.3 Locus Chr. 13 q32 propionyl Coenzyme A carboxylase, beta polypeptide Identifiers Symbol PCCB Entrez 5096 HUGO 8654 OMIM 232050 RefSeq NM_000532 UniProt P05166 Other data EC number 6.4.1.3 Locus Chr. 3 q21-q22 Pathology
A deficiency is associated with propionic acidemia.[15][16][17]
PCC activity is the most sensitive indicator of biotin status tested to date. In future pregnancy studies, the use of lymphocyte PCC activity data should prove valuable in assessment of biotin status.[18]
Regulation
Of Propionyl-CoA Carboxylase
a. Carbamazepine (antiepileptic drug): significantly lowers enzyme levels in the liver[19]
b. E. coli chaperonin proteins groES and groEL: essential for folding and assembly of human PCC heteromeric subunits[20]
c. Bicarbonate: negative cooperativity[21]
d. Mg2+ and MgATP2-: allosteric activation[22]
By Propionyl-CoA Carboxylase
a. 6-Deoxyerythronolide B: decrease in PCC levels lead to increased production [23]
b. Glucokinase in pancreatic beta cells: precursor of beta-PCC shown to decrease KM and increase Vmax; activation [24]
See also
References
- ^ EC 6.4.1.3
- ^ EC 4.1.1.41
- ^ Kalousek F, Darigo MD, Rosenberg LE. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric tetramer of nonidentical subunits. J Biol Chem. 1980 Jan 10;255(1):60-5. PubMed PMID: 6765947
- ^ Diacovich L, Mitchell DL, Pham H, Gago G, Melgar MM, Khosla C, Gramajo H, Tsai SC. Crystal structure of the beta-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochemistry. 2004 Nov 9;43(44):14027-36. PubMed PMID: 15518551
- ^ a b c Huang CS, Sadre-Bazzaz K, Shen Y, Deng B, Zhou ZH, Tong L. Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase. Nature. 2010 Aug 19;466(7309):1001-5. PubMed PMID: 20725044; PubMed Central PMCID: PMC2925307
- ^ Kalousek F, Darigo MD, Rosenberg LE. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric tetramer of nonidentical subunits. J Biol Chem. 1980 Jan 10;255(1):60-5. PubMed PMID: 6765947
- ^ Diacovich L, Mitchell DL, Pham H, Gago G, Melgar MM, Khosla C, Gramajo H, Tsai SC. Crystal structure of the beta-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochemistry. 2004 Nov 9;43(44):14027-36. PubMed PMID: 15518551
- ^ Arabolaza A, Shillito ME, Lin TW, Diacovich L, Melgar M, Pham H, Amick D, Gramajo H, Tsai SC. Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor. Biochemistry. 2010 Aug 31;49(34):7367-76. PubMed PMID: 20690600; PubMed Central PMCID: PMC2927733
- ^ Wolf B, Kalousek F, Rosenberg LE. Essential arginine residues in the active sites of propionyl CoA carboxylase and beta-methylcrotonyl CoA carboxylase. Enzyme. 1979;24(5):302-6. PubMed PMID: 510274
- ^ Sloane V, Waldrop GL. Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for cooperativity in biotin carboxylase. J Biol Chem. 2004 Apr 16;279(16):15772-8. Epub 2004 Feb 11. PubMed PMID: 14960587
- ^ Kalousek F, Darigo MD, Rosenberg LE. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric tetramer of nonidentical subunits. J Biol Chem. 1980 Jan 10;255(1):60-5. PubMed PMID: 6765947
- ^ Hsia YE, Scully KJ, Rosenberg LE. Human propionyl CoA carboxylase: some properties of the partially purified enzyme in fibroblasts from controls and patients with propionic acidemia. Pediatr Res. 1979 Jun;13(6):746-51. PubMed PMID: 481943
- ^ Stubbe J, Fish S, Abeles RH. Are carboxylations involving biotin concerted or nonconcerted? J Biol Chem. 1980 Jan 10;255(1):236-42. PubMed PMID: 7350155
- ^ Reszko AE, Kasumov T, Pierce BA, David F, Hoppel CL, Stanley WC, Des Rosiers C, Brunengraber H. Assessing the reversibility of the anaplerotic reactions of the propionyl-CoA pathway in heart and liver. J Biol Chem. 2003 Sep 12;278(37):34959-65. Epub 2003 Jun 24. PubMed PMID: 12824185
- ^ Ugarte M, Pérez-Cerdá C, Rodríguez-Pombo P, Desviat LR, Pérez B, Richard E, Muro S, Campeau E, Ohura T, Gravel RA (1999). "Overview of mutations in the PCCA and PCCB genes causing propionic acidemia". Hum. Mutat. 14 (4): 275–82. doi:10.1002/(SICI)1098-1004(199910)14:4<275::AID-HUMU1>3.0.CO;2-N. PMID 10502773.
- ^ Desviat LR, Pérez B, Pérez-Cerdá C, Rodríguez-Pombo P, Clavero S, Ugarte M (2004). "Propionic acidemia: mutation update and functional and structural effects of the variant alleles". Mol. Genet. Metab. 83 (1-2): 28–37. doi:10.1016/j.ymgme.2004.08.001. PMID 15464417.
- ^ Deodato F, Boenzi S, Santorelli FM, Dionisi-Vici C (May 2006). "Methylmalonic and propionic aciduria". Am J Med Genet C Semin Med Genet 142C (2): 104–12. doi:10.1002/ajmg.c.30090. PMID 16602092.
- ^ Stratton SL, Bogusiewicz A, Mock MM, Mock NI, Wells AM, Mock DM. Lymphocyte propionyl-CoA carboxylase and its activation by biotin are sensitive indicators of marginal biotin deficiency in humans. Am J Clin Nutr. 2006 Aug;84(2):384-8. PubMed PMID: 16895887; PubMed Central PMCID: PMC1539098
- ^ Rathman SC, Eisenschenk S, McMahon RJ. The abundance and function of biotin-dependent enzymes are reduced in rats chronically administered carbamazepine. J Nutr. 2002 Nov;132(11):3405-10. PubMed PMID: 12421859
- ^ Kelson TL, Ohura T, Kraus JP. Chaperonin-mediated assembly of wild-type and mutant subunits of human propionyl-CoA carboxylase expressed in Escherichia coli. Hum Mol Genet. 1996 Mar;5(3):331-7. PubMed PMID: 8852656
- ^ Sloane V, Waldrop GL. Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for cooperativity in biotin carboxylase. J Biol Chem. 2004 Apr 16;279(16):15772-8. Epub 2004 Feb 11. PubMed PMID: 14960587
- ^ McKeon C, Wolf B. Magnesium and magnesium adenosine triphosphate activation of human propionyl CoA carboxylase and beta-methylcrotonyl CoA carboxylase. Enzyme. 1982;28(1):76-81. PubMed PMID: 6981505
- ^ Zhang H, Boghigian BA, Pfeifer BA. Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol Bioeng. 2010 Feb 15;105(3):567-73. PubMed PMID: 19806677
- ^ Shiraishi A, Yamada Y, Tsuura Y, Fijimoto S, Tsukiyama K, Mukai E, Toyoda Y, Miwa I, Seino Y. A novel glucokinase regulator in pancreatic beta cells: precursor of propionyl-CoA carboxylase beta subunit interacts with glucokinase and augments its activity. J Biol Chem. 2001 Jan 26;276(4):2325-8. Epub 2000 Nov 20. PubMed PMID: 11085976
External links
Ligases: carbon-carbon ligases (EC 6.4) Biotin dependent carboxylase Pyruvate carboxylase · Acetyl-CoA carboxylase · Propionyl-CoA carboxylase · Methylcrotonyl-CoA carboxylaseOther B enzm: 1.1/2/3/4/5/6/7/8/10/11/13/14/15-18, 2.1/2/3/4/5/6/7/8, 2.7.10, 2.7.11-12, 3.1/2/3/4/5/6/7, 3.1.3.48, 3.4.21/22/23/24, 4.1/2/3/4/5/6, 5.1/2/3/4/99, 6.1-3/4/5-6 Synthesis Malonyl-CoA synthesisBeta-ketoacyl-ACP synthase · Β-Ketoacyl ACP reductase · 3-Hydroxyacyl ACP dehydrase · Enoyl ACP reductaseFatty acid desaturasesTriacyl glycerolDegradation Acyl transportGeneralPropionyl-CoA carboxylaseOtherHydroxyacyl-Coenzyme A dehydrogenaseTo acetyl-CoAMetabolism: amino acid metabolism · synthesis and catabolism enzymes (essential in CAPS) K→acetyl-CoA (see below)G G→pyruvate
→citrateG→glutamate→
α-ketoglutarate→alpha-ketoglutarate→TCAOther→succinyl-CoA→TCAG→fumarateasparagine→aspartate→Categories:- Genes on chromosome 13
- Genes on chromosome 3
- EC 6.4.1
Wikimedia Foundation. 2010.
