- Comparative genomic hybridization
-
Comparative genomic hybridization (CGH) or Chromosomal Microarray Analysis (CMA) is a molecular-cytogenetic method for the analysis of copy number changes (gains/losses) in the DNA content of a given subject's DNA and often in tumor cells.
CGH will detect only unbalanced chromosomal changes. Structural chromosome aberrations such as balanced reciprocal translocations or inversions cannot be detected, as they do not change the copy number.
During the 1990s Thomas Cremer realized together with Peter Lichter the concept of comparative genomic hybridization to metaphase chromosomes and to a matrix with DNA spots representing specific genomic sites.
Contents
Method
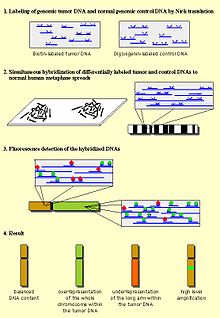
DNA from subject tissue and from normal control tissue (reference) are each labeled with different tags for later analysis by fluorescence. After mixing subject and reference DNA along with unlabeled human cot-1 DNA (placental DNA that is enriched for repetitive DNA sequences such as the Alu and Kpn family)[1] to suppress repetitive DNA sequences, the mix is hybridized to normal metaphase chromosomes or, for array- or matrix-CGH, to a slide containing hundreds or thousands of defined DNA probes. Using epifluorescence microscopy and quantitative image analysis, regional differences in the fluorescence ratio of gains/losses vs. control DNA can be detected and used for identifying abnormal regions in the genome.
Currently, strategically placed Oligos offer a resolution typically of 20–80 base pairs, as compared to the older BAC arrays offering resolution of 100kb.
Uses
Cancer
The method is based on the hybridization of fluorescently labeled tumor DNA (frequently fluorescein (FITC)) and normal DNA (frequently rhodamine or Texas Red) to normal human metaphase preparations.
Other
Aside from analyzing cancer cells, the test is valuable at looking for previously unknown mutations that can lead to children with dysmorphic features, developmental delays, mental retardation, and autism.
Preimplantation genetic diagnosis by CGH may improve the efficiency of IVF in vitro fertilization by increasing embryo implantation rate and reducing multiple pregnancies and spontaneous miscarriages.[2]
Limitations
Chromosomal CGH is capable of detecting loss, gain and amplification of the copy number at the levels of chromosomes. However, it is considered that to detect a single copy loss the region must be at least 5–10 Mb in length. Detection of amplifications (e.g. tens or hundreds of copies of one or few neighboring genes) is known to be sensitive down to less than 1 Mb. Therefore, one must take into consideration that while CGH is sensitive to specific types of copy number gains, the resolution of regional deletions is more limited.
The use of array CGH overcomes many of these limitations, with improvement in resolution and dynamic range, in addition to direct mapping of aberrations to the genome sequence and improved throughput.
Due to the normalization to the most frequent ratio level as "normal", both CGH and array CGH do not provide information as to the ploidy. Since having a balanced DNA content, a tetraploid clone without further rearrangements would appear normal in CGH.
See also
- Array based Comparative Genomic Hybridization
- Oncogene
- Tumor suppressor gene
- Carcinoma
- Sarcoma
- Lymphoma
- Leukemia
- Virtual Karyotype
References
- ^ Invitrogen Corporation --> Human Cot-1 DNA Retrieved on September 9, 2009
- ^ Sher G, Keskintepe L, Keskintepe M, Maassarani G, Tortoriello D, Brody S (January 2009). "Genetic analysis of human embryos by metaphase comparative genomic hybridization (mCGH) improves efficiency of IVF by increasing embryo implantation rate and reducing multiple pregnancies and spontaneous miscarriages". Fertil. Steril. 92 (6): 1886–1894. doi:10.1016/j.fertnstert.2008.11.029. PMID 19135663.
External links
- Virtual Grand Rounds: "Differentiating Microarray Technologies and Related Clinical Implications" by Arthur Beaudet, MD
- Progenetix database: A large collection of CGH and aCGH data from publications (ca. 20000 cases, November 2008).
- CGH article tracker: An attempt to list all (a)CGH publications containing original case data. This is part of the Progenetix project.
- NCBI's Cancer Chromosomes: Cancer Chromosome is an integral part of NCBI's Entrez system, which combines several databases with (molecular-) cytogenetic data.
- A bibliography on copy number variation
Categories:- Molecular genetics
- Gene tests
- Cytogenetics
Wikimedia Foundation. 2010.