- Cyclin D
-
cyclin D1 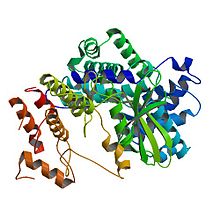
Crystal structure of human cyclin D1 (blue/green) in complex with cyclin-dependent kinase 4 (yellow/red).[1] Identifiers Symbol CCND1 Alt. symbols BCL1, D11S287E, PRAD1 Entrez 595 HUGO 1582 OMIM 168461 RefSeq NM_053056 UniProt P24385 Other data Locus Chr. 11 q13 cyclin D2 Identifiers Symbol CCND2 Entrez 894 HUGO 1583 OMIM 123833 RefSeq NM_001759 UniProt P30279 Other data Locus Chr. 12 p13 cyclin D3 Identifiers Symbol CCND3 Entrez 896 HUGO 1585 OMIM 123834 RefSeq NM_001760 UniProt P30281 Other data Locus Chr. 6 p21 Cyclin D is a member of the cyclin protein family that is involved in regulating cell cycle progression. The synthesis of cyclin D is initiated during G1 and drives the G1/S phase transition. Cyclin D protein is anywhere from 155 (in zebra mussel) to 477 (in Drosophila) amino acids in length.[2]
Contents
Introduction
Once the cells reach a critical cell size (and if no mating partner is present in yeast) and if growth factors and mitogens (for multicellular organism) or nutrients (for unicellular organism) are present, cells enter the cell cycle. In general, all stages of the cell cycle are chronologically separated in humans and are triggered by cyclin-Cdk complexes which are periodically expressed and partially redundant in function. Cyclins are eukaryotic proteins that form holoenzymes with cyclin-dependent protein kinases (Cdk), which they activate. The abundance of cyclins is generally regulated by protein synthesis and degradation through an APC/c dependent pathway.
Cyclin D is one of the major cyclins produced in terms of its functional importance. It interacts with four Cdks: Cdk2, 4, 5, and 6. In proliferating cells, cyclin D-Cdk4/6 complex accumulation is of great importance for cell cycle progression. Namely, cyclin D-Cdk4/6 complex partially phosphorylates Rb, which is able to induce expression of some genes (for example: cyclin E) important for S phase progression.
Mice, Drosophila and many other organisms only have one cyclin D protein. In humans, in addition to the mouse homologue, two more cyclin D proteins have been identified. These human proteins, called cyclin D1, cyclin D2, and cyclin D3 are expressed in most proliferating cells and the relative amounts expressed differs in various cell types.[3]
Homologues
The most studied homologues of cyclin D are found in yeast and viruses.
The yeast homologue of cyclin D, referred to as Cln3, interacts with Cdc28 (cell division control protein) during G1.
In viruses, like Saimiriine herpesvirus 2 (Herpesvirus saimiri) and Human herpesvirus 8 (HHV-8/Kaposi's sarcoma-associated herpesvirus) cyclin D homologues have acquired new functions in order to manipulate the host cell’s metabolism to the virus’ benefit.[4] Viral cyclin D binds human Cdk6 and inhibits Rb by phosphorylating it, which subsequently inhibits expression of genes important for DNA synthesis. Other than Rb, viral cyclin D-Cdk6 complex also targets p27Kip, a Cdk inhibitor of cyclin E and A. In addition, viral cyclin D-Cdk6 is resistant to Cdk inhibitors, such as p21CIP1/WAF1 and p16INK4a which in human cells inhibits Cdk4 by preventing it from forming an active complex with cyclin D.[4] [5]
Function
Cyclins in humans
Growth factors stimulate the Ras/Raf/ERK that induce cyclin D production. One of the members of the pathways, MAPK activates a transcription factor Myc, which alters transcription of genes important in cell cycle, among which is cyclin D. In this way, cyclin D is synthesized as long as the growth factor is present.
Even though cyclin D levels in proliferating cells are sustained as long as the growth factors are present, a key player for G1/S transition is active cyclin D-Cdk4/6 complexes. Despite this, cyclin D has no effect on G1/S transition unless it forms a complex with Cdk 4 or 6.
One of the best known substrates of cyclin D/Cdk4 and -6 is the retinoblastoma tumor suppressor protein (Rb). Rb is an important regulator of genes responsible for progression through the cell cycle, in particular through G1/S phase.
In its un-phosphorylated form, Rb binds a member of E2F family of transcription factors which controls expression of several genes involved in cell cycle progression (example, cyclin E). Rb acts as a repressor, so in complex with E2F it prevents expression of E2F regulates genes, and this inhibits cells from progressing through G1. Active cyclin D/Cdk4 and -6 inhibit Rb by partial phosphorylation, reducing its binding to E2F and thereby allowing E2F-mediated activation of the transcription of the cyclin E gene and the cell progresses towards S-phase. Subsequently, cyclin E fully phosphorylates Rb and completes its inactivation.[6]
Regulation
Regulation in humans
Cyclin D is regulated by the downstream pathway of mitogen receptors via the Ras/MAP kinase and the β-catenin-Tcf/LEF pathways and PI3K. The MAP kinase ERK activates the downstream transcription factors Myc and AP-1 which in turn activate the transcription of the Cdk4, Cdk6 and Cyclin D genes, and increase ribosome biogenesis. Rho family GTPases and Focal Adhesion Kinase (FAK) activate Cyclin D gene in response to integrin.[7]
Members of the cofactor families Cip and Kip, such as p27 and p21, are required for the functionality of the cyclin D Cdk4/6 complex. Surprisingly, Cyclin D is positive regulated by the CKI p27kip. At the same time p27kip inhibits cyclin E and A to prevent a premature entry into S phase and leads to a Cyclin D assembly.
In eukaryotes, overexpression of translation initiation factor 4E (eIF4E) leads to an increased expression levels of cyclin D protein.
Inhibition of cyclin D via i.a. inactivation or degradation leads to an exit of the cell cycle and as well as of differentiation. Inactivation of cyclin D is triggered by several cyclin-dependent kinase inhibitor protein (CKIs) like the INK4 family (e.g. p14, p15, p16, p18). INK4 proteins are activated in response to hyperproliferative stress response that inhibits cell proliferation due to overexpression of e.g. Ras and Myc. Hence, INK4 binds to cyclin D- dependent CDKs and inactivates the whole complex.[3] Glycogen synthase kinase three beta, GSK3β, causes Cyclin D degradation by inhibitory phosphorylation on threonine 286 of the Cyclin D protein. GSK3β is negatively controlled by the PI3K pathway in form of phosphorylation, which is one of several ways in which growth factors regulate cyclin D. Amount of cyclin D in the cell can also be regulated by transcriptional induction, stabilization of the protein, its translocation to the nucleus and its assembly with Cdk4 and Cdk6.[8]
It has been shown that the inhibition of cyclin D (cyclin D1 and 2, in particular) could result from the induction of WAF1/CIP1/p21 protein by PDT. By inhibiting cyclin D, this induction also inhibits Ckd2 and 6. All these processes combined lead to an arrest of the cell in G0/G1 stage.[5]
There are two ways in which DNA damage affects Cdks. Following DNA damage, cyclin D (cyclin D1) is rapidly and transiently degraded by the proteasome. This degradation causes release of p21 from Cdk4 complexes, which inactivates Cdk2 in a p53-independent manner. Another way in which DNA damage targets Cdks is p53-dependent induction of p21, which inhibits cyclin E-Cdk2 complex. In healthy cells, wild-type p53 is quickly degraded by the proteasome. However, DNA damage causes it to accumulate by making it more stable.[3]
Regulation in yeast
A simplification in yeast is that all cyclins bind to the same Cdc subunit, the Cdc28. Cyclins in yeast are controlled by expression, inhibition via CKIs like Far1, and degradation by ubiquitin-mediated proteolysis.[9]
Role in cancer
Given that many human cancers happen in response to errors in cell cycle regulation and in growth factor dependent intracellular pathways, involvement of cyclin D in cell cycle control and growth factor signaling makes it a possible oncogene. In normal cells overproduction of cyclin D shortens the duration of G1 phase only, and considering the importance of cyclin D in growth factor signaling, defects in its regulation could be responsible for absence of growth regulation in cancer cells. Uncontrolled production of cyclin D affects amounts of cyclin D-Cdk4 complex being formed, which can drive the cell through the G0/S checkpoint, even when the growth factors are not present.
Overexpression can happen in one of three ways: as a result of gene amplification, impaired protein degradation, or chromosomal translocation. Gene amplification is responsible for overproduction of cyclin D protein in bladder cancer, espophageal carcinoma, among others.[5]
In cases of sarcomas, colorectal cancers and melanomas, cyclin D overproduction is noted, however, without the amplification of the chromosomal region that encodes it (chromosome 11q13, putative oncogene PRAD1, which has been identified as a translocation event in case of mantle cell lymphoma[10]). In parathyroid adenoma, cyclin D hyper-production is caused by chromosomal translocation, which would place expression of cyclin D (more specifically, cyclin D1) under an inappropriate promoter, leading to overexpression. In this case, cyclin D gene has been translocated to the parathyroid hormone gene, and this event caused abnormal levels of cyclin D.[5] The same mechanisms of overexpression of cyclin D is observed in some tumors of the antibody-producing B cells. Likewise, overexpression of cyclin D protein due to gene translocation is observed in human breast cancer.[5][11]
Additionally, the development of cancer is also enhanced by the fact that retinoblastoma tumor suppressor protein (Rb), one of the key substrates of cyclin D-Cdk 4/6 complex, is quite frequently mutated in human tumors. In its active form, Rb prevents crossing of the G1 checkpoint by blocking transcription of genes responsible for advances in cell cycle. Cyclin D/Cdk4 complex phosphorylates Rb, which inactivates it and allows for the cell to go through the G1. In the event of abnormal inactivation of Rb, in cancer cells, an important regulator of cell cycle progression is lost. When Rb is mutated, levels of cyclin D and p16INK4 are normal.[5]
Another regulator of passage through G1 restriction point is Cdk inhibitor p16, which is encoded by INK4 gene. P16 functions in inactivating cyclin D/Cdk 4 complex. Thus, blocking transcription of INK4 gene would increase cyclin D/Cdk4 activity, which would in turn result in abnormal inactivation of Rb. On the other hand, in case of cyclin D in cancer cells (or loss of p16INK4) wild-type Rb is retained. Due to the importance of p16INK/cyclin D/Cdk4 or 6/Rb pathway in growth factor signaling, mutations in any of the players involved can give rise to cancer.[5]
Mutant phenotype
Studies with mutants suggest that cyclins are positive regulators of cell cycle entry. In yeast, expression of any of the three G1 cyclins triggers cell cycle entry. Since cell cycle progression is related to cell size, mutations in Cyclin D and its homologues show a delay in cell cycle entry and thus, cells with variants in cyclin D have bigger than normal cell size at cell division.[12][13]
p27-/- knockout phenotype show an overproduction of cells because cyclin D is not inhibited anymore, while p27-/- and cyclin D-/- knockouts develop normally.[12]
See also
References
- ^ PDB 2W96; Day PJ, Cleasby A, Tickle IJ, O'Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C, Jhoti H (March 2009). "Crystal structure of human CDK4 in complex with a D-type cyclin". Proc. Natl. Acad. Sci. U.S.A. 106 (11): 4166–70. doi:10.1073/pnas.0809645106. PMC 2657441. PMID 19237565. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2657441.
- ^ http://www.ncbi.nlm.nih.gov/protein?term=cyclin%20D
- ^ a b c Madame Curie Bioscience Database. Eurekah Bioscience Database. http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=eurekah&part=A20842#A20847
- ^ a b Hardwick JM (November 2000). "Cyclin' on the viral path to destruction". Nat. Cell Biol. 2 (11): E203–4. doi:10.1038/35041126. PMID 11056549.
- ^ a b c d e f g Kufe DW, Pollock RE, Weichselbaum RR, Bast RC Ganler TS, Holland JF, Frei E (2003). Cancer medicine 6. Hamilton, Ont: BC Decker. ISBN 1-55009-213-8. http://www.ncbi.nlm.nih.gov/sites/entrez?db=Books&cmd=Search&term=cyc+AND+cmed6[book]&doptcmdl=TOCView&log%24=booksrch&bname=cmed6.
- ^ Resnitzky D, Reed SI (July 1995). "Different roles for cyclins D1 and E in regulation of the G1-to-S transition". Mol. Cell. Biol. 15 (7): 3463–9. PMC 230582. PMID 7791752. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=230582.
- ^ Assoian RK, Klein EA (July 2008). "Growth control by intracellular tension and extracellular stiffness". Trends Cell Biol. 18 (7): 347–52. doi:10.1016/j.tcb.2008.05.002. PMC 2888483. PMID 18514521. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2888483.
- ^ Takahashi-Yanaga F, Sasaguri T (April 2008). "GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy". Cell. Signal. 20 (4): 581–9. doi:10.1016/j.cellsig.2007.10.018. PMID 18023328.
- ^ Bloom J, Cross FR (February 2007). "Multiple levels of cyclin specificity in cell-cycle control". Nat. Rev. Mol. Cell Biol. 8 (2): 149–60. doi:10.1038/nrm2105. PMID 17245415.
- ^ http://www.scbt.com/datasheet-20044-cyclin-d1-dcs-6-antibody.html
- ^ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (1999). Molecular cell biology. New York: Scientific American Books. ISBN 0-7167-3136-3. http://www.ncbi.nlm.nih.gov/sites/entrez?db=Books&cmd=Search&term=cyclin+d+AND+mcb[book]&doptcmdl=TOCView&log%24=booksrch&bname=mcb.
- ^ a b D.H. Sanes, T.A. Reh, W.A. Harris (2005). Development of the Nervous System (2 ed.). Elsevier Ltd, Oxford. ISSN 978-0126186215.
- ^ Geng Y, Yu Q, Sicinska E, Das M, Bronson RT, Sicinski P (January 2001). "Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice". Proc. Natl. Acad. Sci. U.S.A. 98 (1): 194–9. doi:10.1073/pnas.011522998. PMC 14567. PMID 11134518. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=14567.
External links
Cell cycle proteins Cyclin CDK CDK inhibitor P53 p63 p73 family Phases and
checkpointsOther cellular phasesCategories:- Genes on chromosome 11
- Genes on chromosome 12
- Genes on chromosome 6
- Cell cycle
- Proteins
Wikimedia Foundation. 2010.
